Assessment of Microbial Quality of Drinking Water from Different Sources in Selected Districts of Bale Zone, Oromia Regional State, Ethiopia
| Received 02 Mar, 2024 |
Accepted 02 May, 2024 |
Published 03 May, 2024 |
Background and Objective: Safe drinking water is a vital component of human diet. Thus periodic surveillance of the quality of drinking water is critically important for monitoring its safety status. The current study was initiated to assess the microbial quality of drinking water from different sources in the selected districts. Materials and Methods: Microbial analysis of total and faecal coliforms was done using most probable number (MPN) method. Samples were collected following a simple random sampling technique and a cross-sectional study design was employed. Each of the samples were investigated in triplicate. The data were analyzed using descriptive statistics and ANOVA was done at 5% level of probability to investigate the statistical difference among means of contamination between the different sources and localities. Results: A maximum of 6.396±0.010 log10 CFU/mL of SPC has been recorded in water samples from Tagona River in Goba District. The highest faecal coliform and Escherichia coli contamination of 1101 and 972.800±128.200 MPN/100 mL were recorded in rivers Bamo and Tagona in Goba and Haro Wanji of Dello Mana and Bamo River, respectively, showing similar trends across the three agro-ecologies. Rivers and ponds were categorized under high to very high risk classifications. Conclusion: Thus, society, as an emergency action, should take actions like filtration, boiling and treatment with some commercially available antimicrobial agents. The government is expected to establish facilities for the supply of safe drinking water to avoid the likely health burden to be posed on society from the highly contaminated water sources.
INTRODUCTION
Sustainability of life is ensured by the accessibility of safe drinking water1 and satisfactory supply should be available to all2. Safe drinking water is recognized as that having no remarkable health threat to consumers. The use of multiple barriers should be planned carefully from catchment to point of use to ensure prevention of microbial as well as physical contamination3. World Health Organization4,5 reported that more than 80% of human diseases in the World are attributed to the use of unsafe drinking water or inadequate sanitary practices. Cholera, shigellosis and campylobacteriosis are among the important waterborne diseases easily contracted due to contaminated water sources serving. Recent WHO report briefed that about 1.1 billion people, out of which 42%6, globally drink unsafe water and the vast majority of diarrhoeal diseases in the World, estimated to be nearly 88%, are attributed to unsafe drinking water, sanitation and hygiene. The consumption of unsafe water is responsible for 3.1% (1.7 million), of the annual deaths and 3.7% (54.2 million) of the annual health burden. In general, most waterborne outbreaks involve source contamination, breakdown of the treatment system, contamination of the distribution system and the use of untreated water7.
It is estimated that about 75% of the health problems in Ethiopia are communicable arising from inaccessibility to safe and adequate water supply8. In the country, the number of companies engaged in production and trading of packaged/bottled water is increasing from time to time9. The Ethiopian water quality standard requires that no E. coli or thermoduric bacteria and coliform bacteria are detected in the treated water entering the distribution system and water in the distribution system10. Bacteriological analysis of drinking water (tap, spring and well) from around Dire Dawa City showed that all the samples (100%) from spring and tap were positive for indicator microorganisms (total coliforms, thermotolerant (faecal) coliforms. Whereas 50% of the tap water samples were found to be contaminated with the same organisms11. Assessment of the microbiological quality of drinking water in four districts of Addis Ababa City reported that 6% of tap water, 6% of reservoir samples and 24% of spring samples were contaminated with bacteria12. Likewise, microbial quality study of drinking water in Debre Zeyit Town found that 100% of the drinking water samples from underground sources were contaminated with total coliforms and 20% was found to be positive for faecal coliforms and faecal streptococci. Thus, regular examination of the drinking water resource with regard to microbiological and physicochemical quality is of crucial importance so that appropriate remedial actions can be forwarded to safeguard the community.
So far, quality of drinking water from different sources has not been investigated in Bale Zone in terms of microbiology. Therefore, this study was initiated to assess the microbiological quality of drinking water from different sources across the different agro-ecologies of Bale Zone of Oromia Regional State, Ethiopia.
MATERIALS AND METHODS
Sample collection: Drinking water samples were collected from purposively selected Districts of Bale Zone, namely, Goro, Goba and Dallo Manna, Oromia Regional State in consultation with Regional Bureau, Zonal and District Offices of Water Resources. The assessments were carried out on samples from three selected districts of the zones. A total of 135 samples of drinking water were collected randomly from different sources in the zone. Analyses were done in plant pathology laboratory of Sinana Agricultural Research Center in late October 2021 which is the last month of the heavy rainy season in the Zone.
Microbial analysis: Microrobial analysis for the determination of total coliforms, faecal coliforms and E. coli was performed employing most probable number (MPN) technique6.
Further identification of coliforms was done by carrying out the appropriate biochemical tests like, indole production, methyl red, voges/proskauer, citrate utilization, motility, gram staining and gas production from lactose following techniques used by Arbab et al.8.
Statistical analysis: All the microbial count data were subjected to descriptive analysis and means, minimum and maximum values and standard error were calculated using SPSS 20. Moreover, Analysis of Variance (ANOVA) was also performed using SAS 9.1.3 to investigate the significance of variability among the values obtained from different sources and agro-ecologies at 5% level of probability. Significantly different means were separated using Duncan Multiple Range Test (DMRT).
RESULTS AND DISCUSSION
Total bacterial count: All drinking water samples collected from the 9 kebeles of the three districts of Bale Zone from different sources (Fig. 1) were tested for their hygienic status in terms of total bacterial count. In Goro District, a total of 45 samples were collected out of which 42 were from tap and only 3 were from unprotected spring. There was statistically non significant variability between the kebeles of the district, with the samples from spring in Waltai Gobu scoring the highest log10 CFU/mL of 6.132 and the minimum value of 6.009 log10 CFU/mL was obtained from tapped drinking water from the same district (Table 1). Water samples from Garre and Chaffe Mana kebeles were non significantly different from those of Waltai Gobu.
A relatively higher log10 CFU/mL was recorded from samples collected from Goba District. The maximum count (6.396 log10 CFU/mL) was from samples of Tagona River in Waltai Sura kebele, whereas the least record (6.159) was from samples collected from households in Waltai Tosha kebele that fetched the water from Bamo River.
 
|
| Table 1: | Total bacterial counts (log10 CFU/mL) for drinking water samples obtained from different sources across the study districts | |||
| District | Kebele | Drinking water source | N | Minimum | Maximum | Mean | p<0.05 |
| Goro | Waltai Gobu | Tapped | 12 | 5.538 | 6.244 | 6.009±0.068cd | <0.0001 |
| Spring | 3 | 5.925 | 6.239 | 6.132±0.104c | 0.0003 | ||
| Garre | Tapped | 15 | 5.621 | 6.244 | 6.014±0.050cd | <0.0001 | |
| Chafe Mana | Tapped | 15 | 5.834 | 6.436 | 6.026±0.044cd | <0.0001 | |
| Goba | Waltai Sura | River (Tagona) | 4 | 6.368 | 6.41 | 6.396±0.010c | <0.0001 |
| River (Tagona-household) | 4 | 6.301 | 6.417 | 6.349±0.024c | <0.0001 | ||
| River (Micha-household) | 5 | 6.073 | 6.394 | 6.245±0.064c | <0.0001 | ||
| Bore hole (Burgullo) | 2 | 6.354 | 6.382 | 6.368±0.014c | 0.001 | ||
| Waltai Tosha | River (Bamo) | 8 | 5.837 | 6.428 | 6.307±0.057cde | <0.0001 | |
| River (Bamo-household) | 7 | 6.135 | 6.436 | 6.159±0.080c | <0.0001 | ||
| Aloshe | Hand pump | 13 | 6.089 | 6.401 | 6.238±0.037c | <0.0001 | |
| Bore hole | 2 | 6.089 | 6.401 | 6.231±0.097c | 0.218 | ||
| Dallo Manna | Haya Oda | River (Yadot-household) | 6 | 5.842 | 5.851 | 5.847±0.004cde | <0.0005 |
| River (Erba) | 4 | 5.374 | 5.718 | 5.516±0.104de | <0.0005 | ||
| River (Erba-household) | 5 | 5.189 | 5.64 | 5.384±0.095e | <0.0004 | ||
| Barraq | Tapped | 4 | 8.307 | 8.362 | 8.330±0.013a | <0.0001 | |
| Pond (Haro Sora) | 8 | 7.675 | 8.36 | 8.080±0.077ab | <0.0001 | ||
| Pond (Haro Wanji) | 3 | 7.658 | 8.293 | 8.012±0.187ab | 0.0005 | ||
| Gongoma | Tapped | 4 | 6.403 | 8.348 | 7.810±0.469b | 0.0005 | |
| Spring | 2 | 8.26 | 8.35 | 8.305±0.045ab | 0.0034 | ||
| River (Gongoma) | 9 | 7.882 | 8.393 | 8.204±0.081ab | 0.0001 | ||
| Means with different letter are significantly different at p<0.05 and Mean±Standard Error | |||||||
Dallo Manna samples were collected from 4 sources (river (53%, tap 18%, pond 24% and spring 4%). Samples from tap collected from Barraq kebele exhibited the highest level of bacterial contamination with log10 CFU/mL of 8.330. Samples from spring, Gongoma River and Haro Sora pond took the 2nd, 3rd and 4th places in bacterial contamination with values of 8.305, 8.204 and 8.080 log10 CFU/mL, respectively (Table 1).
Total coliform count: A highly significant difference was observed in total coliform count among the drinking water samples of Goro District. Samples from spring at Waltai Gobu and tap water from Chafe Mana kebeles had the least coliform contamination of 32.663 and 60.661 MPN/100 mL, respectively, of water with non-significant statistical variation. On the contrary, a higher total coliform count of 95 CFU/100 mL was recorded from tap water samples in Nekemte Town13. However, tapped water from Garre kebele had coliform contamination of 549.933 MPN/100 mL (Table 2). A lower value ranging from 1.50±0.71 CFU/100 mL to 133.67±21.25 CFU/100 mL was reported by Amenu et al.11, from unprotected well and tap water samples in Dire Dawa Administrative Council. A similarly lower level of contamination was reported in drinking water samples from spring (2-70 MPN/100 mL) and hand pipe (2-9 MPN/100 mL) by Negera et al.14, in a study conducted in Shashemene rural districts. In Fiche town still, lower total coliform count range of 3.93 to 9.29 CFU/mL was recorded in dry and wet seasons, respectively from samples of piped drinking water15.
Total coliform counts have been shown to radically increase in samples from Goba District. The test showed that samples from Tagona River (Waltai Sura), Bamo River (Waltai Tosha), hand pump and unprotected borehole (Aloshe) showed a coliform count range of 1101.0 to 1023.54 MPN/100 mL of sample. This finding agreed with the result of Helmi et al.16, reporting a total coliform range of 270 to 1600 MPN/100 mL of drinking water samples from river. Similarly, a total coliform count range of 67 to 1366 MPN/100 mL of sample drinking water from wells. Household drinking water samples (rivers and unprotected bore hole) from Waltai Sura kebele had a cell number range of 670.000 to 756.000 MPN/100 mL (Table 2).
In Dallo Manna District, out of the 9 sources used for sampling, drinking water samples from the 7 sources had a total coliform cell count of 885 MPN/100 mL. Only samples of tap from Barraq kebele scored a relatively lower count of 593.75 MPN/100 mL of sample (Table 2). The analysis result showed that none of the samples comply with the WHO guideline7.
| Table 2: | Total coliform counts (log10 CFU/mL) for drinking water samples obtained from different sources across the study districts | |||
| District | Kebele | Drinking water source | N | Minimum | Maximum | Mean | p<0.05 | Detection (%) |
| Goro | Waltai Gobu | Tapped | 12 | 2.99 | 1101 | 368.999±128.661cd | 0.015 | 91.7 |
| Spring | 3 | 2.99 | 75 | 32.663±21.730d | 0.272 | 66.7 | ||
| Garre | Tapped | 15 | 5.621 | 6.244 | 549.933±124.432bc | 0.001 | 100 | |
| Chafe Mana | Tapped | 15 | 2.99 | 150 | 60.661±17.978d | 0.005 | 46.7 | |
| Goba | Waltai Sura | River (Tagona) | 4 | 1101 | 1101 | 1101.00±0.000a | 0 | 100 |
| River (Tagona-household) | 4 | 240 | 1101 | 670.500±248.549abc | 0.074 | 100 | ||
| River (Micha-household) | 5 | 240 | 1101 | 756.600±210.901abc | 0.023 | 100 | ||
| Bore hole (Burgullo) | 2 | 240 | 1100 | 670.000±0.014abc | 0.3632 | 100 | ||
| Waltai Tosha | River (Bamo) | 8 | 240 | 1101 | 1014.90±86.100ab | <0.0001 | 100 | |
| River (Bamo-household) | 7 | 1101 | 1101 | 1101.00±0.000a | 0 | 100 | ||
| Aloshe | Hand pump | 13 | 95 | 1101 | 1023.54±77.378ab | <0.0001 | 100 | |
| Bore hole | 2 | 1100 | 1101 | 1100.50±0.500a | 0.0003 | 100 | ||
| Dallo Manna | Haya Oda | River (Yadot-household) | 6 | 5.988 | 5.988 | 1100.67±0.333a | 0 | 100 |
| River (Erba) | 4 | 240 | 1101 | 885.750±215.250ab | 0.026 | 100 | ||
| River (Erba-household) | 5 | 1100 | 1101 | 1100.80±0.200a | <0.0001 | 100 | ||
| Barraq | Tapped | 4 | 23 | 1101 | 593.750±294.006abc | <0.0001 | 100 | |
| Pond (Haro Sora) | 8 | 1101 | 1101 | 1101.00±0.000a | 0 | 100 | ||
| Pond (Haro Wanji) | 3 | 1101 | 1101 | 1101.00±0.000a | 0 | 100 | ||
| Gongoma | Tapped | 4 | 1101 | 1101 | 1101.00±0.000a | 0 | 100 | |
| Spring | 2 | 1101 | 1101 | 1101.00±0.000a | 0 | 100 | ||
| River (Gongoma) | 9 | 1101 | 1101 | 1101.00±0.000a | 0 | 100 | ||
| Means with different letter are significantly different at p<0.05 and Mean±Standard Error | ||||||||
Fecal coliform count: A maximum faecal coliform count of 107.183 MPN/100 mL was obtained from tested tap water samples of Waltai Gobu kebele. Samples from spring in the same kebele had the lowest faecal coliform count of 9.197 MPN/100 mL. Whereas tap water samples from Garre and Chafe Mana kebeles had shown a faecal coliform contamination of 26.075 and 56.693 MPN/100 mL, respectively (Table 3). Amenu et al.11, reported that all water samples were found to be contaminated by faecal coliforms.
On the other hand, tests of water samples obtained directly from Tagona and Bamo Rivers and households in Waltai Sura and Waltai Tosha kebeles, respectively, who depend on the two rivers for drinking water indicated that the highest MPN of 1101 and above was obtained. Whereas, the highest faecal coliform value of 54 CFU/100 mL was reported from protected well11. Samples collected from hand pump and bore hole in Aloshe kebele showed the lowest faecal coliform contamination of 14.677 and 2.99 MPN, respectively (Table 3). But the lowest value of faecal coliform (0.34 CFU/100 mL) was obtained from tap water samples in Dire Dawa.
There was a significant statistical difference between the sources in faecal coliform count in Dallo Manna District with a range stretching from 14.1 to 1101.0 MPN across the three kebeles. Tap water samples from Gongoma and Barraq recorded lowered faecal coliform counts of 14.050 and 21.000 MPN (Table 3). Samples from Haro Sora Pond, Erba river and Haro Wanji Pond were found to have 725.125, 972.600 and 1100.67 MPN, respectively.
In general, according to risk classification for thermotolerant coliforms or E. coli for rural water supplies cited by Ashuro et al.17, drinking water sources such as Bamo and Tagona Rivers at Goba District, Erba and Yadot Rivers at Delo Mana, some piped supplies in Goro District, ponds like Haro Wanji and Haro Sora at Dallo Mana have been found to be classified under “high” to “very high” risk categories. On the contrary, only samples from borehole in Aloshe kebele of Goba District seemed to conform with WHO guidelines6.
| Table 3: | Faecal coliform counts (log10 CFU/mL) for drinking water samples obtained from different sources across the study districts | |||
| District | Kebele | Drinking water source | N | Minimum | Maximum | Mean | p<0.05 | Risk category |
| Goro | Waltai Gobu | Tapped | 12 | 2.99 | 1100 | 107.183±90.343ef | 0.2605 | HR |
| Spring | 3 | 2.99 | 21 | 9.197±5.904f | 0.2596 | LR | ||
| Garre | Tapped | 15 | 7.3 | 240 | 59.693±15.962f | 0.0022 | IR | |
| Chafe Mana | Tapped | 15 | 2.99 | 210 | 26.075±13.609f | 0.076 | IR | |
| Goba | Waltai Sura | River (Tagona) | 4 | 1100 | 1101 | 1100.75±0.250a | <0.0001 | VHR |
| River (Tagona-household) | 4 | 1101 | 1101 | 1101.00±0.000a | 0 | VHR | ||
| River (Micha-household) | 5 | 43 | 1101 | 515.600±241.172cd | 0.0993 | HR | ||
| Bore hole (Burgullo) | 2 | 460 | 460 | 460.000±0.000cde | 0 | HR | ||
| Waltai Tosha | River (Bamo) | 8 | 1100 | 1101 | 1100.80±0.133a | 0 | VHR | |
| River (Bamo-household) | 7 | 1101 | 1101 | 1101.00±0.000a | 0 | VHR | ||
| Aloshe | Hand pump | 13 | 2.99 | 150 | 14.677±11.282f | 0.2177 | IR | |
| Bore hole | 2 | 2.99 | 2.99 | 2.990±0.000f | 0 | IC | ||
| Dallo Manna | Haya Oda | River (Yadot-household) | 6 | 93 | 210 | 151.000±33.778def | 0.0466 | HR |
| River (Erba) | 4 | 93 | 1100 | 633.250±271.144bc | 0.1016 | HR | ||
| River (Erba-household) | 5 | 460 | 1101 | 972.600±128.150ab | 0.0016 | HR | ||
| Barraq | Tapped | 4 | 15 | 23 | 21.000±2.000f | 0.0018 | IR | |
| Pond (Haro Sora) | 8 | 240 | 1101 | 725.125±144.800abc | 0.0016 | HR | ||
| Pond (Haro Wanji) | 3 | 1100 | 1101 | 1100.67±0.333a | <.0001 | VHR | ||
| Gongoma | Tapped | 4 | 9.1 | 23 | 14.050±3.292f | 0.0236 | IR | |
| Spring | 2 | 20 | 210 | 115.000±95.000ef | 0.4396 | HR | ||
| River (Gongoma) | 9 | 11 | 1101 | 356.778±148.387cdef | 0.0429 | HR | ||
| IC: In conformity with WHO guidelines, LR: Low risk, IR: Intermediate risk, HR: High risk, VHR: Very high risk, means with different letter are significantly different at p<0.05 and Mean±Standard Error | ||||||||
Escherichia coli count: Samples from each of the different sources were also tested for the detection of E. coli. It is in the spring water samples that the lowest MPN/100 mL of E. coli cells (8.863) was recorded in Waltai Gobu kebele of Goro District. Tap water samples from the same kebele had an E. coli count value of 12.098 CFU/100 mL. With non-significant variability, 11.055 CFU/100 mL were counted in samples from the similar source from Chafe Mana District. The maximum MPN/100 mL (42.607) was recorded in tap water samples from Garre District (Table 4). A comparably lower E. coli value of 6.0±0.54 was reported in Kenya in tap water samples18.
However, in Goba District, the highest MPN of E. coli of 972.800 was obtained from samples collected from households that fetched the water from Bamo river for drinking. Unprotected borehole (Burgullo) and Bamo River samples took the 2nd (780.500) and the 3rd (696.800) places in terms of MPN of E. coli. A similar report showed that the highest contamination level of 160.0±14.14 CFU/mL was detected in rainwater samples in a study published in Kenya18. Water samples with almost no E. coli (2.99 cells/100 mL) were obtained from a protected bore hole in Aloshe kebele of the district (Table 4). Similarly, an equivalent value of 3.554 cells was recoded in samples from hand pumps in the same kebele. Drinking water samples form Tagona and Micha Rivers (both household and source) showed relatively lower MPN of E. coli ranging from 328.5 to 514.0 (Table 4).
Water samples from Erba River drinking households were found to host the highest number of E. coli cells (884.400) per 100 mL. However, samples directly taken from the river showed a significantly lower MPN of E. coli (398.250). At Barraq kebele, samples from Haro Wanji pond and tap showed a lower E. coli cell population of 12.663 and 20.000 per 100 mL, respectively. Gongoma and Yadot Rivers exhibited an MPN of 60.566 and 84.333, in Gongoma and Haya Oda kebeles, respectively (Table 4).
According to risk classification for thermotolerant coliforms or E.coli for rural water supplies cited by Ashuro et al.17, all the rivers in Goba being used as sources of drinking water for the local society were under “high risk” category. Similarly, Erba River, Haro Sora pond and spring at Dallo Mana have fallen under “high risk” classification (Table 4). Bore hole sourced drinking water have conformed with WHO guideline. Samples from spring in Goro and hand pump in Goba were under “low risk category”. Almost none of the samples have complied with WHO guidelines and Ethiopian standards for drinking water quality19 except that of bore hole in Goba District.
|
| Table 4: | Escherichia coli counts (log10 CFU/mL) for drinking water samples obtained from different sources across the study districts | |||
| District | Kebele | Drinking water source | N | Minimum | Maximum | Mean | p (<0.05) | Risk category |
| Goro | Waltai Gobu | Tapped | 12 | 2.99 | 43 | 12.098±3.227g | 0.0032 | IR |
| Spring | 3 | 2.99 | 20 | 8.863±5.571g | 0.2526 | LR | ||
| Garre | Tapped | 15 | 6.2 | 240 | 42.607±19.329fg | 0.0447 | IR | |
| Chafe Mana | Tapped | 15 | 2.99 | 43 | 11.0553±3.571g | 0.0079 | IR | |
| Goba | Waltai Sura | River (Tagona) | 4 | 28 | 1101 | 328.500±259.0143defg | 0.2942 | HR |
| River (Tagona-household) | 4 | 35 | 1101 | 514.000±219.818bcde | 0.1014 | HR | ||
| River (Micha-household) | 5 | 3.6 | 1101 | 443.660±268.360cdef | 0.1736 | HR | ||
| Bore hole (Burgullo) | 2 | 460 | 1101 | 780.500±320.500abc | 0.2481 | HR | ||
| Waltai Tosha | River (Bamo) | 8 | 93 | 1101 | 696.800±139.014abcd | 0.0007 | HR | |
| River (Bamo-household) | 7 | 460 | 1101 | 972.800±128.200a | 0.0016 | HR | ||
| Aloshe | Hand pump | 13 | 2.99 | 9.1 | 3.554±0.466g | <.0001 | LR | |
| Bore hole | 2 | 2.99 | 2.99 | 2.990±0.000g | 0 | IC | ||
| Dallo Manna | Haya Oda | River (Yadot-household) | 6 | 15 | 210 | 84.333±62.945fg | 0.3123 | IR |
| River (Erba) | 4 | 93 | 1100 | 398.250±237.896cdefg | 0.1927 | HR | ||
| River (Erba-household) | 5 | 20 | 1101 | 884.400±216.100ab | 0.0149 | HR | ||
| Barraq | Tapped | 4 | 11 | 23 | 20.000±3.000g | 0.0069 | IR | |
| Pond (Haro Sora) | 8 | 35 | 460 | 267.500±63.259efg | 0.0039 | HR | ||
| Pond (Haro Wanji) | 3 | 2.99 | 20 | 12.663±5.047g | 0.1289 | IR | ||
| Gongoma | Tapped | 4 | 9.1 | 23 | 12.575±3.475g | 0.0363 | IR | |
| Spring | 2 | 11 | 210 | 110.500±99.500fg | 0.4667 | HR | ||
| River (Gongoma) | 9 | 2.99 | 210 | 60.566±28.388fg | 0.0654 | IR | ||
| IC: In conformity with WHO guidelines, LR: Low risk, IR: Intermediate risk, HR: High risk, VHR: Very high risk, means with different letter are significantly different at p<0.05 and Mean±Standard Error | ||||||||
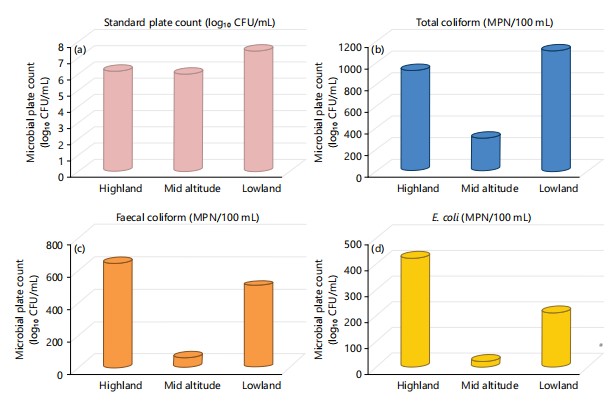
Microbial contamination across the different agro-ecologies: Microbial contamination of the drinking water samples looks different for the different agro ecologies across the zone. Standard plate count showed an increasing trend with decreasing altitude (Fig. 2a). However, total coliform was lower at mid altitude as compared to high and low lands with still being highest at lowlands (Fig. 2b). Feacal coliform contamination of drinking water followed a opposite looking trend with the highest contamination being at the highland agro ecologies. Similar to that of coliforms, the lowest population was observed at the mid altitude districts (Fig. 2c). Similarly, E. coli distribution followed the same trend as that of faecal coliforms with the highest level being highlands and the second pick being observed at lowlands and the least reported at mid altitude areas (Fig. 2d).
RECOMMENDATION
Thus, the society, as an emergency action, should take actions like filtration, boiling and treatment with some commercially available antimicrobial agents following manufacturers’ instructions. Furthermore, the water sector should take an immediate action in performing appropriate treatments (chlorination) at the source/reservoirs for piped distribution. In addition, the distribution lines should be periodically inspected and maintained for proper functionality. For majority of our farmers/pastoralists who depend on polluted river and highly turbid ponds for drinking water, the sector should make great endeavor to establish facilities for the supply of safe drinking water to avoid the likely health burden to be posed on the society from the highly contaminated water sources.
CONCLUSION
Most of the samples from river are seriously contaminated by total coliform, faecal coliform and E. coli showing high sewage discharge and disposal of urban waste into the rivers. High microbial contamination of fenced ponds are most probably from animal dung carried on foot steps of those fetching water. Moreover, inadequate periodic maintenance of the entire distribution system led to detection of total coliforms in tap water samples making it riskfull for consumption.
SIGNIFICANCE STATEMENT
This study was initiated to assess the microbiological quality of drinking water samples taken from different sources. The investigation made it clear, for the first time that local people including those living surrounding towns and the rural people were consuming drinking water from highly dirty rivers into which urban wastes and household trash were being thrown and not protected totally. Nearly all the samples from the different sources didn’t comply with the national and international microbiological standards hence classified as high risk. Thus, this study brought into the knowledge and attention of the scientific society and the policy makers, the possible health hazard the society is suffering from due to consumption of highly contaminated drinking water from those sources.
ACKNOWLEDGMENT
We are sincerely grateful to Oromia Agricultural Research Institute for supporting the research project in financial (grant number: IQQO/FS/FM-2017 (1)) and technical terms.
REFERENCES
- Alhassan, H. and P.A. Kwakwa, 2014. When water is scarce: The perception of water quality and effects on the vulnerable. J. Water Sanit. Hyg. Dev., 4: 43-50.
- WHO, 2003. The World Health Report 2003 Shaping the Future. World Health Organization, Geneva, Switzerland, ISBN: 9789241562430, Pages: 193.
- WHO, 2011. Guidelines for Drinking-Water Quality. 4th Edn., World Health Organization, Geneva, Switzerland, ISBN-13: 9789241548151, Pages: 564.
- WHO, 2008. Guidelines for Drinking-Water Quality: 3rd Edition: Volume 1-Recommendations Incorporating the First and Second Addenda. 3rd Edn., World Health Organization, Geneva, Switzerland, ISBN: 9789241547611, Pages: 668.
- WHO and UNICEF., 2010. Progress on Sanitation and Drinking-Water. World Health Organization and United Nations Children's Fund, Geneva, Switzerland, ISBN: 9789241563956, Pages: 60.
- WHO, 1996. Guidelines For Drinking-Water Quality, 2nd Edition: Volume 2-Health Criteria and Other Supporting Information. 2nd Edn., World Health Organization, Geneva, Switzerland, ISBN: 9241544805, Pages: 990.
- WHO, 2017. Guidelines for Drinking-Water Quality, 4th edition, Incorporating the 1st Addendum. 4th Edn., World Health Organization, Geneva, Switzerland, ISBN: 978-92-4-154995-0, Pages: 631.
- Arbab, S., H. Ullah, W. Wang, K. Li, A. Akbar and J. Zhang, 2021. Isolation and identification of infection-causing bacteria in dairy animals and determination of their antibiogram. J. Food Qual., 2021.
- Ensermu, M., 2014. Trends in bottled water use survey in Addis Ababa: Implication on reverse logistics of bottled water manufacturing in Ethiopia. Int. J. Sci. Res., 3: 934-942.
- Debebe, D., Z. Getaneh and F. Behulu, 2022. Bacterial contamination of school’s drinking water in Addis Ababa, Ethiopia. Zede J., 40: 23-32.
- Amenu, D., S. Menkir and T. Gobena, 2013. Microbiological quality of drinking water sources and water handling practices among rural communities of Dire Dawa Administrative Council. Int. J. Curr. Res. Acad. Rev., 1: 29-54.
- Wolde, A.M., K. Jemal, G.M. Woldearegay and K.D. Tullu, 2020. Quality and safety of municipal drinking water in Addis Ababa City, Ethiopia. Environ. Health Prev. Med., 25.
- Duressa, G., F. Assefa and M. Jida, 2019. Assessment of bacteriological and physicochemical quality of drinking water from source to household tap connection in Nekemte, Oromia, Ethiopia. J. Environ. Public Health, 2019.
- Negera, E., G. Nuro and M. Kebede, 2017. Microbiological assessment of drinking water with reference to diarrheagenic bacterial pathogens in Shashemane Rural District, Ethiopia. Afr. J. Microbiol. Res., 11: 254-263.
- Sebsibe, I., B. Degaga, G. Feye and T. Tekle, 2021. Bacteriological and physical quality of fiche drinking water from households and reservoirs, Oromia, Ethiopia. Water Pract. Technol., 16: 924-934.
- Helmi, K., F. Barthod, G. Méheut, A. Henry, F. Poty, F. Laurent and N. Charni-Ben-Tabassi, 2015. Methods for microbiological quality assessment in drinking water: A comparative study. J. Water Health, 13: 34-41.
- Ashuro, Z., M.B. Aregu, G.G. Kanno, B. Negassa and N.E. Soboksa et al., 2021. Bacteriological quality of drinking water and associated factors at the internally displaced people sites, Gedeo Zone, Southern Ethiopia: A cross-sectional study. Environ. Health Insights, 15.
- Onyango, A.E., M.W. Okoth, C.N. Kunyanga and B.O. Aliwa, 2018. Microbiological quality and contamination level of water sources in Isiolo County in Kenya. J. Environ. Public Health, 2018.
- Lewoyehu, M., 2021. Evaluation of drinking water quality in rural area of Amhara Region, Ethiopia: The case of Mecha District. J. Chem., 2021.
How to Cite this paper?
APA-7 Style
Tegegn,
A., Negesso,
Z., Oluma,
T. (2024). Assessment of Microbial Quality of Drinking Water from Different Sources in Selected Districts of Bale Zone, Oromia Regional State, Ethiopia. Asian Journal of Emerging Research, 6(1), 13-21. https://doi.org/10.3923/ajer.2024.13.21
ACS Style
Tegegn,
A.; Negesso,
Z.; Oluma,
T. Assessment of Microbial Quality of Drinking Water from Different Sources in Selected Districts of Bale Zone, Oromia Regional State, Ethiopia. Asian J. Emerg. Res 2024, 6, 13-21. https://doi.org/10.3923/ajer.2024.13.21
AMA Style
Tegegn
A, Negesso
Z, Oluma
T. Assessment of Microbial Quality of Drinking Water from Different Sources in Selected Districts of Bale Zone, Oromia Regional State, Ethiopia. Asian Journal of Emerging Research. 2024; 6(1): 13-21. https://doi.org/10.3923/ajer.2024.13.21
Chicago/Turabian Style
Tegegn, Addisu, Zeneba Negesso, and Tolera Oluma.
2024. "Assessment of Microbial Quality of Drinking Water from Different Sources in Selected Districts of Bale Zone, Oromia Regional State, Ethiopia" Asian Journal of Emerging Research 6, no. 1: 13-21. https://doi.org/10.3923/ajer.2024.13.21

This work is licensed under a Creative Commons Attribution 4.0 International License.



